Filter Messages by Concept Type
In this example, we will filter patient messages by concept type (concept_type) in a couple of different ways. These code samples use version 2.0.0 of the ScienceIO API. Before you begin, please check your version and upgrade your scienceio Python package if necessary.
To see a more basic filtering example, go to Analytics with Pandas page.
For this exercise only, remember that:
- When
concept_typeequals “Medical Conditions”, theconcept_nameindicates the specific side effect. - When
concept_typeequals “Chemicals & Drugs”, theconcept_nameindicates the medication.
Option 1: Get a Count of Side Effects
One way to examine trends in messages is to use counts. After running the code from Structure, Parse, and Analyze Messages, add this code to get a count of all side effects that were identified in the messages.
# Subset the df to only medical conditions
side_effects_db = structured_results[structured_results["concept_type"]=="Medical Conditions"]
# Create a count of each
side_effects_db["concept_name"].value_counts()
The resulting table looks like this:
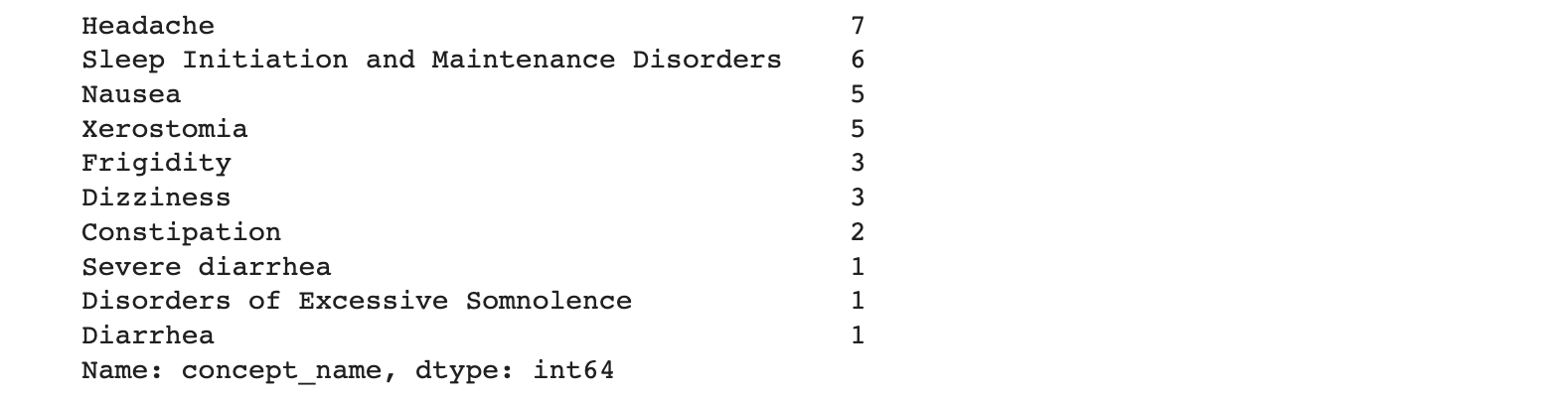
Notes:
- The name of the referenced DataFrame in this example is
structured_results, which we created in step 3 of Structure, Parse, and Analyze Messages. - The
concept_typeequals “Medical Conditions” because the API assigned all side effects the Medical Conditions concept type when it structured the messages. - The
concept_nameis what generates the count for each side effect; in this example, the API assigned all side effects to theconcept_namevariable when it structured the messages.
concept_type you want to count. ScienceIO has nine different concept types.Option 2: Filter by Medications
Another way to examine messages is to list every concept_name found for a particular concept_type. After running the code from Structure, Parse, and Analyze Messages, add this code to create a list of all medications that were identified in the messages.
# Subset the df to only medications
structured_results[structured_results["concept_type"]=="Chemicals & Drugs"]
The resulting table looks like this:

Notes:
- The name of the referenced DataFrame in this example is
structured_results, which we created in step 3 of Structure, Parse, and Analyze Messages. - The
concept_typeequals “Chemicals & Drugs” because the API assigned all medications the Chemicals & Drugs concept type when it structured the messages.
concept_type you want to list. ScienceIO has nine different concept types.Option 3: Find Side Effects Associated with Medications
Continuing on from Options 1 and 2 above, meta-analyses are possible to help provide a deeper understanding of your data.
Important
This example is based on the simplifying assumption that each patient message mentions only a single medication. The patient messages in this example also included only two different concept types, and both apply to this meta-analysis (so we did not filter out any others). You may need to first filter by concept type when trying the code on your own dataFirst, use this code to create a mapping of the medication to each patient message. This will add another column called medication to the DataFrame, which indicates the medication a patient mentioned in their message.
structured_results_medications = structured_results.copy()
medication_map = {}
for _,row in structured_results_medications.iterrows():
if row["concept_type"]=="Chemicals & Drugs":
medication_map[row["message_id"]] = row["concept_name"]
print(medication_map)
#
structured_results_medications["medication"] = structured_results_medications["message_id"].apply(lambda x: medication_map[x])
structured_results_medications.head()
The results look like this:
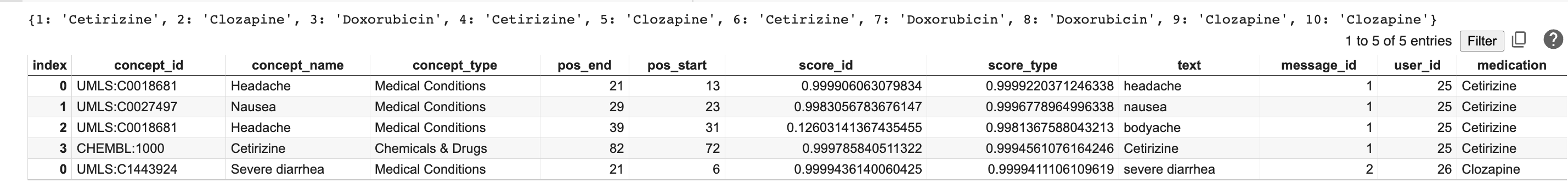
Next, use pandas to tie each medication to its reported side effect by filtering the structured results DataFrame to only the “Medical Conditions” concept_type, grouping the results by the new medication column, and adding a count in the concept_name column.
# Filter to only medical conditions
filtered_conditions = structured_results_medications[structured_results_medications["concept_type"]=="Medical Conditions"]
filtered_conditions.groupby("medication")["concept_name"].value_counts().to_frame("counts")
The resulting table looks like this:

Questions?
If you need additional help, we’re standing by ready to assist! Contact support.
Feedback
Was this page helpful?
Great! If you ever have questions or want to provide feedback, send us an email.
Bummer. We hate when we miss the mark. If you have suggestions for improvements or other general comments, send us an email.