Integrate with Chroma
Using ScienceIO with Chroma allows you to capture and semantically search on a large database of unstructured clinical documents, enabling efficient answering of healthcare questions. In this example, we’ll use the ScienceIO embeddings endpoint to extract embeddings from healthcare text documents, store that encoded information in a Chroma collection, and create a vector database. Then, we’ll query the database to find a specific text string from the original healthcare text.
Step 1: Setup
Install Chroma and ScienceIO
Use this code if you do not have Chroma and ScienceIO installed on your machine, or if you are starting a new Jupyter notebook. Otherwise, skip to Import Packages.
# Only required if not already installed
# Use !pip install if you receive a SyntaxError
pip install chromadb
pip install scienceio
Troubleshooting
If you are working in a Juypter notebook and receive a SyntaxError, use!pip install as the syntax instead.Import Packages
Next, import everything you will need to successfully run the code. These include:
DocumentsandEmbeddingsfrom Chromapandas(under the pd alias)- the
ScienceIOlibrary
# Import required packages
import chromadb
from chromadb.api.types import Documents, Embeddings
import pandas as pd
from scienceio import ScienceIO
Step 2: Get Embeddings and Create a Chroma Database
Next, we’ll call the ScienceIO embeddings endpoint and use the results to create a vector database built from a Chroma collection.
Define a Variable to Hold Your Text Documents
The list_of_docs variable contains your input text to process for embeddings (this example uses three different text strings as the input text). Each separate entity of input text/paragraph(s) is called a “document” by Chroma.
[] should be enclosed in parenthesis and separated with a comma. This is where you can swap out the sample data for your own real text documents.# Define the variable to hold the list of input text strings
# Swap out your own real text documents here
list_of_docs = ["This genomic report provides information on the patient's tumor molecular profile and may inform the selection of targeted therapies and clinical trial opportunities. Further molecular testing, including validation and dynamic monitoring, may be necessary to guide personalized treatment decisions.",
"Administer Androgen Deprivation Therapy as ordered, Administer pain management as needed,Consult with urologist for further management and treatment options, Consult with dietitian for nutrition plan, Consult with physical therapist for mobility and exercise plan",
"This blood work laboratory report provides results of various tests performed on a blood sample collected from the patient. The tests were performed using standard laboratory methods and equipment. TEST RESULTS: Hematology: Red Blood Cells (RBC): 4.5 million/µL (reference range: 4.5 - 5.5 million/µL)"
]
Create a Custom Function for Embeddings
Define a custom function that calls the embeddings endpoint. This function will take the document(s) in the list_of_docs variable, call the endpoint, and return the embeddings.
# Define the custom function to create embeddings using ScienceIO's API
def scienceio_embed_function(texts: Documents) -> Embeddings:
# Call the Embeddings endpoint and embed the documents
scio = ScienceIO()
embeddings = []
for text in texts:
response = scio.embeddings(text)
embeddings.append(response['embeddings'])
return embeddings
Create the Chroma Collection for the Database
Get the Chroma client, create a collection, and add the text documents and embeddings to it.
# Create and define the Chroma collection for the db
def create_chroma_db(documents, name, embedding_function):
chroma_client = chromadb.Client()
db = chroma_client.create_collection(name=name, embedding_function=embedding_function)
for i,d in enumerate(documents):
db.add(
documents=d,
ids=str(i)
)
return db
Build the Database
Build a database using the Chroma collection, and confirm that the embeddings were captured for each document and are working properly.
# Build a db using the Chroma collection
db = create_chroma_db(list_of_docs, "scienceio_chroma_db",scienceio_embed_function)
# Confirm the documents were embedded properly
pd.DataFrame(db.peek(3))
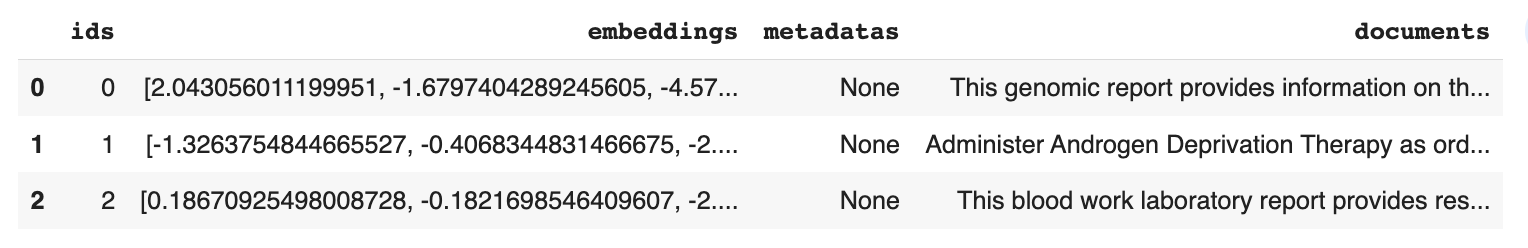
You should see this:
Step 3: Search for a Specific Text String
Set up a function that allows you to semantically query the collection to find a specific text string, and only return the top result (n_results=1). In this example, we are looking for “red blood cells.” Feel free to search for a different string.
# Define a function to query the db and return the top result
def get_relevant_passage(query, db):
passage = db.query(query_texts=[query], n_results=1)['documents'][0][0]
return passage
# Query for the text "red blood cells"
result = get_relevant_passage("red blood cells", db)
result
The result looks like this:
This blood work laboratory report provides results of various tests performed on a blood sample collected from the patient. The tests were performed using standard laboratory methods and equipment. TEST RESULTS: Hematology: Red Blood Cells (RBC): 4.5 million/µL (reference range: 4.5 - 5.5 million/µL)
Feedback
Was this page helpful?
Great! If you ever have questions or want to provide feedback, send us an email.
Bummer. We hate when we miss the mark. If you have suggestions for improvements or other general comments, send us an email.