Retrieval Augmented Generation (RAG) for Medical Records Using ScienceIO + OpenAI
In this example, we’ll safely leverage OpenAI to query a database of medical records that contains protected health information (PHI). In order to minimize exposure of the PHI, we’ll first use ScienceIO’s redact_phi endpoint to scrub the medical records of identified PHI. Then, we’ll use the embeddings endpoint to create a vector database of the redacted records, which can be safely read by OpenAI when we perform the query.
Important
You must have an API Key from OpenAI to successfully run this example. You will insert that API key in step 5.Step 1: Setup
Install Chroma, OpenAI, and ScienceIO
Use this code if you do not have these packages installed on your machine, or if you are starting a new Jupyter notebook. Otherwise, skip to Import Necessary Packages.
# Only required if not already installed
# Use !pip install if you receive a SyntaxError
pip install chromadb
pip install openai
pip install scienceio
Troubleshooting
If you are working in a Jupyter notebook and receive a SyntaxError, use!pip install as the syntax instead.Import Necessary Packages
Next, import all of the packages you will need to successfully run the code. These include:
chromadb(the Chroma database)- the
DocumentsandEmbeddingscollections from Chroma - the
ScienceIOlibrary pandas(under the pd alias)openai(the OpenAI client library)
# Import required packages
import chromadb
from chromadb.api.types import Documents, Embeddings
from scienceio import ScienceIO
import pandas as pd
import openai
Step 2: Load the Medical Records
For this example, we’ll use some medical records for a patient that contain dummy PHI. We’ll load a mock intake form, laboratory report, genomic report, and pathology report into a docs variable. This variable contains two key:value pairs: the doc_type key indicates the type of record, and the doc_text key contains the record text.
Feel free to use your own medical records containing real PHI.
# Load medical records into a variable
docs = [
{'doc_type': "genomic_report",
'doc_text': """PATIENT ID: 123456
PATIENT NAME: Adalberto Q Patterson
DATE OF TESTING: 02/03/2023
INTRODUCTION:
This report summarizes the genomic analysis of a patient with a suspected solid tumor, performed using Foundation Medicine's comprehensive genomic profiling (CGP) assay.
METHODS:
DNA was extracted from the patient's formalin-fixed, paraffin-embedded (FFPE) tissue sample.
Next-generation sequencing (NGS) was performed on the extracted DNA to generate over 400 million reads per sample.
The sequencing data was analyzed to identify genetic alterations, including single nucleotide variations (SNVs), insertions and deletions (indels), copy number alterations (CNAs), and structural variants (SVs).
RESULTS:
The genomic analysis revealed the presence of the following genomic alterations:
PIK3CA mutation (c.1322G>A)
TP53 mutation (c.217G>T)
ERBB2 amplification
CDK12 deletion
These genomic alterations are commonly observed in various types of solid tumors and may contribute to the patient's tumorigenesis and potential therapeutic targets.
CONCLUSIONS:
This genomic analysis provides a comprehensive and in-depth understanding of the patient's tumor and may inform treatment options, including targeted therapy and clinical trials. Further studies, including validation of these findings and assessment of additional genomic markers, may be needed to guide patient management.
PATHOLOGIST:
Joana Jannette, MD, Board Certified Pathologist.
"""},
{'doc_type':'intake_form',
'doc_text':"""Patient Information:
Name: Adalberto Q Patterson
Age: 60 years
Gender: Male
Date of Arrival: 2023-02-03
Time of Arrival: 14:00 PM
Contact Information:
Address: 331 Embers Trace St, Red Devil, OH, USA 02602
Phone: (613) 456-3156
Reason for Visit:
Severe lower abdominal pain and urinary urgency
Medical History:
Current Diagnosis: Prostate Cancer
Allergies: None reported
ECOG Score: 0
Medications:
a. Androgen Deprivation Therapy: Leuprolide 7.5 mg every 4 weeks
b. Pain Management: Acetaminophen 500 mg orally every 6 hours as needed
Previous Hospitalizations: None reported
Family History:
a. Mother: Died of lung cancer at age 70
b. Father: Died of heart disease at age 75
c. Siblings: One brother, alive and well
Social History:
a. Smoking: Former smoker, quit 20 years ago
b. Alcohol Use: Occasionally drinks beer
c. Substance Abuse: None reported
d. Sexual History: Heterosexual, monogamous relationship with spouse
e. Occupation: Retired
f. Support System: Married, two children, supportive spouse
Vital Signs:
Blood Pressure: 140/90 mmHg
Heart Rate: 70 bpm
Respiratory Rate: 18 breaths/minute
Oxygen Saturation: 98% on room air
Temperature: 98.7°F
Physical Exam:
General Appearance: Well-developed, well-nourished male in moderate distress
Head, Eyes, Ears, Nose, Throat: No abnormalities noted
Chest and Lungs: Clear to auscultation bilaterally
Cardiovascular: Regular rhythm, no murmurs or rubs
Abdomen: Soft, non-tender, non-distended
Extremities: No swelling, no edema
Genitourinary: Tenderness and swelling noted in right lower quadrant, positive for CVA tenderness
Skin: Warm, dry, intact
Diagnostic Tests:
Radiology: Abdominal/pelvic CT scan ordered
Pathology: Prostate biopsy ordered
Treatment Plan:
Administer Androgen Deprivation Therapy as ordered
Administer pain management as needed
Consult with urologist for further management and treatment options
Consult with dietitian for nutrition plan
Consult with physical therapist for mobility and exercise plan
Signatures:
Patient Signature: _________________________
Caregiver Signature: ________________________
Physician Signature: ________________________
"""},
{'doc_type':'lab_report',
'doc_text':"""PATIENT ID: 123456
PATIENT NAME: Adalberto Q Patterson
DATE OF SAMPLE COLLECTION: 01/01/2023
INTRODUCTION:
This blood work lab report summarizes the results of various laboratory tests performed on the patient's blood sample. The tests were performed to evaluate various aspects of the patient's health, including blood cell count, liver function, kidney function, and electrolyte balance.
TEST RESULTS:
Complete Blood Count (CBC):
White Blood Cell (WBC) count: 7.0 x 10^3/uL (normal range: 4.0 - 11.0)
Red Blood Cell (RBC) count: 4.5 x 10^6/uL (normal range: 4.0 - 5.0)
Hemoglobin (Hb) concentration: 14.5 g/dL (normal range: 12.0 - 15.5)
Hematocrit (Hct) value: 43.0% (normal range: 37.0 - 47.0)
Platelet (Plt) count: 250 x 10^3/uL (normal range: 150 - 450)
Liver Function Tests:
Aspartate Aminotransferase (AST) level: 25 U/L (normal range: 0 - 40)
Alanine Aminotransferase (ALT) level: 30 U/L (normal range: 0 - 44)
Alkaline Phosphatase (ALP) level: 100 U/L (normal range: 40 - 129)
Bilirubin (total) level: 1.2 mg/dL (normal range: 0.3 - 1.2)
Kidney Function Tests:
Blood Urea Nitrogen (BUN) level: 12 mg/dL (normal range: 6 - 20)
Creatinine (Cr) level: 0.7 mg/dL (normal range: 0.6 - 1.3)
Creatinine Clearance (CrCl) value: 106 mL/min (normal range: 88 - 128)
Electrolyte Tests:
Sodium (Na) level: 136 mEq/L (normal range: 135 - 145)
Potassium (K) level: 4.5 mEq/L (normal range: 3.5 - 5.0)
Chloride (Cl) level: 100 mEq/L (normal range: 98 - 106)
CONCLUSION:
The patient's laboratory test results are generally within normal ranges and do not suggest any significant abnormalities. However, some of the results, such as the slightly elevated liver function test levels, may indicate the need for further evaluation and testing.
REPORT GENERATED BY:
ABC Laboratories
Laboratory Director: Salome Valk, PhD, D(ABMLI)
"""},
{'doc_type':'pathology_report',
'doc_text':"""Patient Name: Adalberto Q Patterson
Patient ID: 1234567
Date of Report: 2023-02-03
Microscopic Examination:
Specimen type: Biopsy
Diagnosis: Malignant melanoma
The biopsy shows evidence of malignant melanoma, a type of skin cancer that occurs when the pigment-producing cells (melanocytes) become malignant. The tissue is characterized by the presence of atypical melanocytes with marked nuclear pleomorphism, hyperchromasia and abundant cytoplasm. The cells form nests and clusters, and are frequently surrounded by a pigmented host response. These findings are consistent with a diagnosis of malignant melanoma.
Immunohistochemistry:
Melan-A, S100: Positive
HMB-45: Positive
The results of the immunohistochemical stains are consistent with the diagnosis of malignant melanoma. Melan-A and S100 are markers of melanocytes and were both positive in the biopsy specimen. HMB-45 is a more specific marker for malignant melanoma and was also positive in the tissue.
Comment:
This biopsy confirms the diagnosis of malignant melanoma and appropriate clinical management, including surgical excision and possible additional treatment, should be initiated. Further studies, including imaging, are recommended to determine the extent of disease.
Pathologist: Dr. Sheba Brandt
Signature: [Signature not included in text]
"""}]
Step 3: Redact the PHI
Next, we’ll call the redact_phi endpoint and redact identified PHI from the medical records. This action will scrub the identified PHI before sending a record to OpenAI during a query. Each redacted record will be stored within the docs variable inside a new key:value pair called doc_redacted.
# Create a scio object
scio = ScienceIO()
# Loop through each note and call the redact_phi endpoint
redacted_responses = []
for text in docs:
response = scio.redact_phi(text["doc_text"])
text.update({'doc_redacted': response['output_text']})
# Print the original and redacted medical records
docs[0]
The result looks like this:
{'doc_type': 'genomic_report',
'doc_text': "PATIENT ID: 123456\nPATIENT NAME: Adalberto Q Patterson\nDATE OF TESTING: 02/03/2023\n\nINTRODUCTION:\nThis report summarizes the genomic analysis of a patient with a suspected solid tumor, performed using Foundation Medicine's comprehensive genomic profiling (CGP) assay.\n\nMETHODS:\n\nDNA was extracted from the patient's formalin-fixed, paraffin-embedded (FFPE) tissue sample.\nNext-generation sequencing (NGS) was performed on the extracted DNA to generate over 400 million reads per sample.\nThe sequencing data was analyzed to identify genetic alterations, including single nucleotide variations (SNVs), insertions and deletions (indels), copy number alterations (CNAs), and structural variants (SVs).\nRESULTS:\n\nThe genomic analysis revealed the presence of the following genomic alterations:\nPIK3CA mutation (c.1322G>A)\nTP53 mutation (c.217G>T)\nERBB2 amplification\nCDK12 deletion\nThese genomic alterations are commonly observed in various types of solid tumors and may contribute to the patient's tumorigenesis and potential therapeutic targets.\nCONCLUSIONS:\nThis genomic analysis provides a comprehensive and in-depth understanding of the patient's tumor and may inform treatment options, including targeted therapy and clinical trials. Further studies, including validation of these findings and assessment of additional genomic markers, may be needed to guide patient management.\n\nPATHOLOGIST:\nJoana Jannette, MD, Board Certified Pathologist.\n",
'doc_redacted': "PATIENT ID: [MEDICALRECORD]\nPATIENT NAME: [PATIENT]\nDATE OF TESTING: [DATE]\n\nINTRODUCTION:\nThis report summarizes the genomic analysis of a patient with a suspected solid tumor, performed using Foundation Medicine's comprehensive genomic profiling (CGP) assay.\n\nMETHODS:\n\nDNA was extracted from the patient's formalin-fixed, paraffin-embedded (FFPE) tissue sample.\nNext-generation sequencing (NGS) was performed on the extracted DNA to generate over 400 million reads per sample.\nThe sequencing data was analyzed to identify genetic alterations, including single nucleotide variations (SNVs), insertions and deletions (indels), copy number alterations (CNAs), and structural variants (SVs).\nRESULTS:\n\nThe genomic analysis revealed the presence of the following genomic alterations:\nPIK3CA mutation (c.1322G>A)\nTP53 mutation (c.217G>T)\nERBB2 amplification\nCDK12 deletion\nThese genomic alterations are commonly observed in various types of solid tumors and may contribute to the patient's tumorigenesis and potential therapeutic targets.\nCONCLUSIONS:\nThis genomic analysis provides a comprehensive and in-depth understanding of the patient's tumor and may inform treatment options, including targeted therapy and clinical trials. Further studies, including validation of these findings and assessment of additional genomic markers, may be needed to guide patient management.\n\nPATHOLOGIST:\n[DOCTOR], MD, Board Certified Pathologist.\n"}
Step 4: Transform the Text Into a Vector Database
Using ScienceIO’s embeddings with Chroma allows you to capture and semantically search on a large database of unstructured clinical documents, enabling efficient answering of healthcare questions. Before we query the redacted medical records, we’ll need to create two custom functions to allow us to build the vector database:
- The first function takes the document(s) in the
docsvariable, calls theembeddingsendpoint, and returns the embeddings. - The second function creates a Chroma collection, then adds the documents and ScienceIO embeddings to it. It also captures the
doc_typeas metadata.
Once we’ve done this, we can build the vector database using the Chroma collection, and preview the first few lines to confirm the database looks as expected.
# Define a custom function to create embeddings
def scienceio_embed_function(texts: Documents) -> Embeddings:
# Call the embeddings endpoint and embed the documents
scio = ScienceIO()
embeddings = []
for text in texts:
response = scio.embeddings(text)
embeddings.append(response['embeddings'])
return embeddings
# Create and define the Chroma collection for the db
def create_chroma_db(documents, name, embedding_function):
chroma_client = chromadb.Client()
db = chroma_client.get_or_create_collection(name=name, embedding_function=embedding_function)
for i,d in enumerate(documents):
db.add(
documents=d['doc_redacted'],
ids=str(i),
metadatas=[{'doc_type':d["doc_type"]}]
)
return db
# Build a db using the Chroma collection
db = create_chroma_db(docs, "redacted_notes",scienceio_embed_function)
# Preview the first few lines of the db
pd.DataFrame(db.peek(4))
The result looks like this:
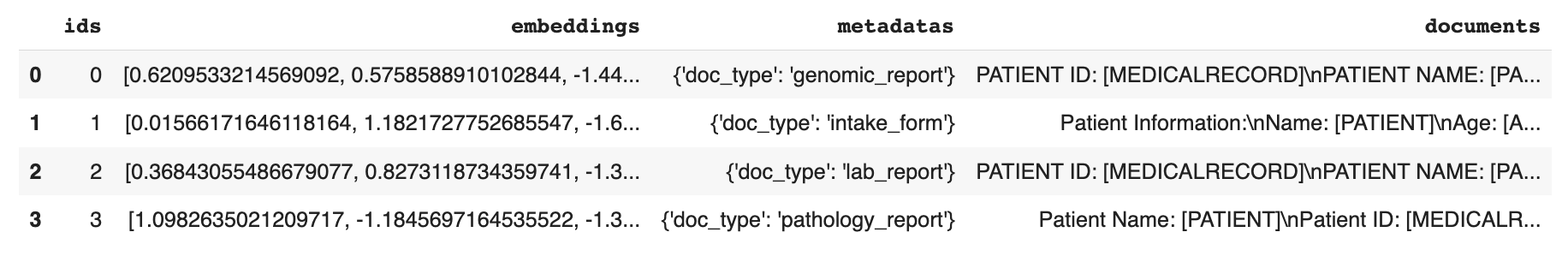
Step 5: Define the Custom Functions for the Query
Now, we’ll define a custom function to query the database by searching for the vector that is most similar to the input query, based on cosine similarity or other similarity measures. Then, we’ll define a second custom function to use GPT-3.5 for text generation of the answer based on the redacted record that was found during the query.
Note that your functions will not do anything until you add query text that leverages them, as shown in Step 6.
openai.api_key = "<INSERT YOUR OPENAI API KEY HERE>"It should look like:
openai.api_key = "ab-cdEFGHIjKLmNopqRS12TU3VwxyZAAbbCcDde4EFFggHHIi5"# Define a custom function to query the db and return the top result
def get_relevant_document(query, db):
passage = db.query(query_texts=[query], n_results=1)
if passage is None:
return None
else:
return passage
# Define a custom function to generate a text answer from a document using OpenAI
# Be sure to add your OPENAI API key where indicated
def generate_answer_from_doc(question:str,doc:str) -> str:
openai.api_key = "<INSERT YOUR OPENAI API KEY HERE>"
response = openai.Completion.create(
engine="text-davinci-003",
prompt=f"{doc}\n{question}",
max_tokens=150
)
answer = response['choices'][0]['text'].strip()
# Return the answer
return answer
Step 6: Query the Database
Now, we can leverage the custom functions from Step 5 to run some queries on the medical records. To do this, add some additional code (two examples are given below) that allows you to provide the query text while also leveraging the get_relevant_document and generate_answer_from_doc functions. We’ll also get the metadata so that we can see the doc_type.
You can swap out your own question using either of the examples below.
Note
You may be subject to rate limits from OpenAI, depending on your plan. If you receive aRateLimitError, check your billing plan on OpenAI and/or contact them for assistance.Example 1: Query for Lab Tests Related to Kidneys
In this example, we’ll ask the question, “What are the results of the kidney-related laboratory tests that were performed?”
# Query for lab tests related to kidneys
question = "What are the results of the kidney-related laboratory tests that were performed?"
document = get_relevant_document(question, db)
if document is not None:
answer = generate_answer_from_doc(question=question, doc=document['documents'][0][0])
print(f"Answer to the question:\n{question}\nis:")
print("-"*100)
print(answer)
print("-"*100)
print(f"Relevant metadata is {document['metadatas'][0][0]}.\nDocument text is:")
print(document['documents'][0][0])
else:
print("No relevant document found")
The response looks like this:
Answer to the question:
What are the results of the kidney-related laboratory tests that were performed?
is:
----------------------------------------------------------------------------------------------------
The results of the kidney-related laboratory tests that were performed were as follows: Blood Urea Nitrogen (BUN) level: 12 mg/dL (normal range: 6 - 20); Creatinine (Cr) level: 0.7 mg/dL (normal range: 0.6 - 1.3); Creatinine Clearance (CrCl) value: 106 mL/min (normal range: 88 - 128).
----------------------------------------------------------------------------------------------------
Relevant metadata is {'doc_type': 'lab_report'}.
Document text is:
PATIENT ID: [MEDICALRECORD]
PATIENT NAME: [PATIENT]
DATE OF SAMPLE COLLECTION: [DATE]
INTRODUCTION:
This blood work lab report summarizes the results of various laboratory tests performed on the patient's blood sample. The tests were performed to evaluate various aspects of the patient's health, including blood cell count, liver function, kidney function, and electrolyte balance.
TEST RESULTS:
Complete Blood Count (CBC):
White Blood Cell (WBC) count: 7.0 x 10^3/uL (normal range: 4.0 - 11.0)
Red Blood Cell (RBC) count: 4.5 x 10^6/uL (normal range: 4.0 - 5.0)
Hemoglobin (Hb) concentration: 14.5 g/dL (normal range: 12.0 - 15.5)
Hematocrit (Hct) value: 43.0% (normal range: 37.0 - 47.0)
Platelet (Plt) count: 250 x 10^3/uL (normal range: 150 - 450)
Liver Function Tests:
Aspartate Aminotransferase (AST) level: 25 U/L (normal range: 0 - 40)
Alanine Aminotransferase (ALT) level: 30 U/L (normal range: 0 - 44)
Alkaline Phosphatase (ALP) level: 100 U/L (normal range: 40 - 129)
Bilirubin (total) level: 1.2 mg/dL (normal range: 0.3 - 1.2)
Kidney Function Tests:
Blood Urea Nitrogen (BUN) level: 12 mg/dL (normal range: 6 - 20)
Creatinine (Cr) level: 0.7 mg/dL (normal range: 0.6 - 1.3)
Creatinine Clearance (CrCl) value: 106 mL/min (normal range: 88 - 128)
Electrolyte Tests:
Sodium (Na) level: 136 mEq/L (normal range: 135 - 145)
Potassium (K) level: 4.5 mEq/L (normal range: 3.5 - 5.0)
Chloride (Cl) level: 100 mEq/L (normal range: 98 - 106)
CONCLUSION:
The patient's laboratory test results are generally within normal ranges and do not suggest any significant abnormalities. However, some of the results, such as the slightly elevated liver function test levels, may indicate the need for further evaluation and testing.
REPORT GENERATED BY:
[ORGANIZATION]
Laboratory Director: [DOCTOR], PhD, D([HOSPITAL])
Example 2: Query for Genomic Alterations
In this example, we’ll ask the question, “What genomic alterations were detected in the patient’s genomic testing reports?”
# Query for genomic alterations
question = "What genomic alterations were detected in the patient's genomic testing reports?"
document = get_relevant_document(question, db)
if document is not None:
answer = generate_answer_from_doc(question=question, doc=document['documents'][0][0])
print(f"Answer to the question:\n{question}\nis:")
print("-"*100)
print(answer)
print("-"*100)
print(f"Relevant metadata is {document['metadatas'][0][0]}.\nDocument text is:")
print(document['documents'][0][0])
else:
print("No relevant document found")
The response looks like this:
Answer to the question:
What genomic alterations were detected in the patient's genomic testing reports?
is:
----------------------------------------------------------------------------------------------------
The genomic analysis revealed the presence of the following genomic alterations: PIK3CA mutation (c.1322G>A), TP53 mutation (c.217G>T), ERBB2 amplification, and CDK12 deletion. These alterations are commonly observed in various types of solid tumors and may contribute to the patient's tumorigenesis and potential therapeutic targets.
----------------------------------------------------------------------------------------------------
Relevant metadata is {'doc_type': 'genomic_report'}.
Document text is:
PATIENT ID: [MEDICALRECORD]
PATIENT NAME: [PATIENT]
DATE OF TESTING: [DATE]
INTRODUCTION:
This report summarizes the genomic analysis of a patient with a suspected solid tumor, performed using Foundation Medicine's comprehensive genomic profiling (CGP) assay.
METHODS:
DNA was extracted from the patient's formalin-fixed, paraffin-embedded (FFPE) tissue sample.
Next-generation sequencing (NGS) was performed on the extracted DNA to generate over 400 million reads per sample.
The sequencing data was analyzed to identify genetic alterations, including single nucleotide variations (SNVs), insertions and deletions (indels), copy number alterations (CNAs), and structural variants (SVs).
RESULTS:
The genomic analysis revealed the presence of the following genomic alterations:
PIK3CA mutation (c.1322G>A)
TP53 mutation (c.217G>T)
ERBB2 amplification
CDK12 deletion
These genomic alterations are commonly observed in various types of solid tumors and may contribute to the patient's tumorigenesis and potential therapeutic targets.
CONCLUSIONS:
This genomic analysis provides a comprehensive and in-depth understanding of the patient's tumor and may inform treatment options, including targeted therapy and clinical trials. Further studies, including validation of these findings and assessment of additional genomic markers, may be needed to guide patient management.
PATHOLOGIST:
[DOCTOR], MD, Board Certified Pathologist.
Feedback
Was this page helpful?
Great! If you ever have questions or want to provide feedback, send us an email.
Bummer. We hate when we miss the mark. If you have suggestions for improvements or other general comments, send us an email.