Structure and Filter Clinical Notes
In this example, we’ll structure a clinical note with the structure endpoint, and then use pandas DataFrames and Python functions to filter the structured data out or in based on various concepts or keywords. We’ll also include the location of our filtered data so that we know where each concept or keyword was mentioned within the original clinical note.
Step 1: Import Packages
First, import and set up all of the packages you will need to successfully run the code. These include:
- the
ScienceIOlibraryscio = ScienceIO()creates thescioobject
pandas(under the pd alias)pd.set_optionis used to set the column width for the resulting table
- the
jsonpackage - the
Listcollection type from thetypingPython module
# Import and set up required packages
from scienceio import ScienceIO
scio = ScienceIO()
import pandas as pd
pd.set_option('display.max_colwidth', 0)
import json
from typing import List
Troubleshooting
If you are getting a ModuleNotFoundError, make sure you have installed the scienceio Python Package within the tool and/or notebook you are using. You may do this via the UI of some editors by searching for and adding the scienceio package, or by using thepip install scienceio command within the notebook.Step 2: Call the Endpoint
Next, call the structure endpoint using an unstructured clinical note. This allows you to structure all of the clinical note text so that it can be used in further analysis.
# Call the `structure` endpoint with an unstructured clinical note
query_text = """
Name: Jane Dutton
Age: 24 years old
Gender: Female
Medical History:
Hypertension (diagnosed 2 years ago)
Oral Thrush (recently diagnosed)
Tattoos (no known complications)
Clinical Note:
Jane Doe is a 24-year-old female patient who presented with symptoms of oral thrush. She reports experiencing difficulty swallowing and a persistent white coating on her tongue. Upon examination, a white, curd-like material was observed on the tongue and oral mucosa. The diagnosis of oral thrush was made, and treatment with antifungal medication was prescribed.
The patient's medical history includes a diagnosis of hypertension, which was made 5 years ago. She reports being compliant with her medication regimen and reports no recent changes in her blood pressure readings. No significant abnormalities were observed on physical examination.
The patient also reports having multiple tattoos, with no known complications. She reports that she received all her tattoos from licensed and reputable tattoo artists and had no allergic reactions or infections after receiving them. However, the patient was advised to continue practicing proper wound care and hygiene to prevent any potential infections.
The patient was educated on the importance of maintaining good oral hygiene to prevent the recurrence of oral thrush. She was also advised to continue taking her hypertension medications as prescribed and to monitor her blood pressure regularly.
The patient was scheduled for a follow-up visit in two weeks to monitor her progress with the treatment for oral thrush and to check her blood pressure readings.
"""
response = scio.structure(query_text)
print(json.dumps(response,indent=4))
The resulting response includes spans that break out every healthcare concept identified. Here is the first span you will see (full response not shown due to length):
"spans": [
{
"concept_id": "UMLS:C0001779",
"concept_name": "Age",
"concept_type": "Anatomy & Physiology",
"pos_end": 24,
"pos_start": 21,
"score_id": 0.9999144077301025,
"score_type": 0.9999768733978271,
"text": "Age"
},
Step 3: Create a DataFrame from the Results
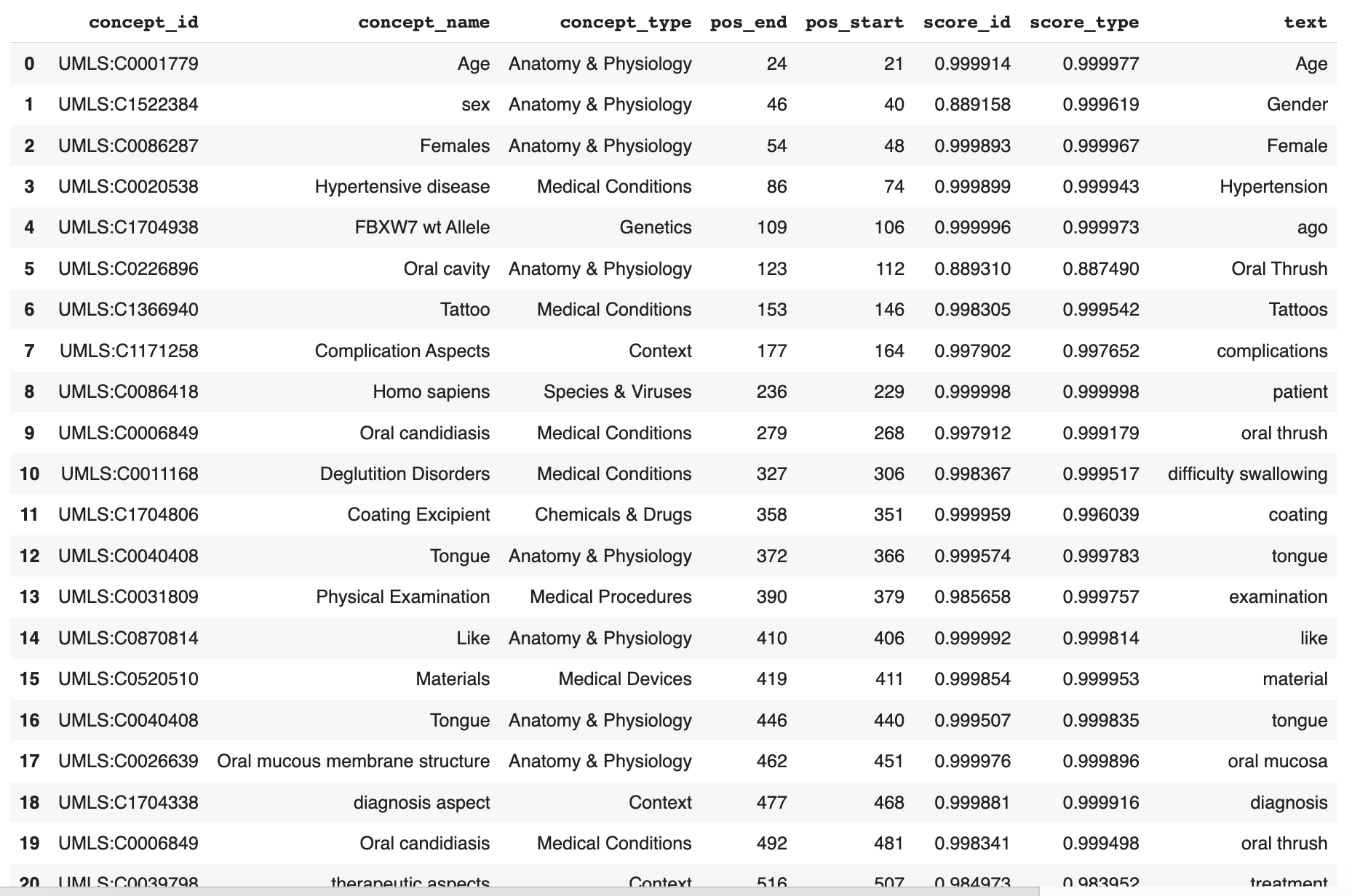
The next step is to decide which data to filter in or out to make our analysis more meaningful. Do this by looking through the JSON response to find each concept_id, concept_name, concept_type, or just text string that you’d like to filter in or out. To make this process easier, create a pandas DataFrame from the JSON response so that you can easily browse the concepts and keywords. This DataFrame will also be used as the basis for the Python functions in Step 4.
# Create a pandas df from results
df = pd.DataFrame(response["spans"])
df
Step 4: Define the Concepts or Keywords to Filter
As mentioned above, you can filter based on any concept (concept_id, concept_name, concept_type) or text string you identify in the JSON response. These examples offer a few options for you to try.
Option 1: Filter By UMLS Codes
Use Case: I want to filter out the UMLS codes I know I’m not interested in.
ScienceIO assigns every piece of healthcare text a unique identifier based on the primary ontology (in this example, a UMLS code), so there may be structured text (like “Homo sapiens”) that you know you are not interested in. Filtering out the UMLS code tied to this type of text will create a smaller DataFrame that can be better analyzed.
In this option, we’ll define a generic_concepts_list variable that contains each UMLS code (taken from the concept_id in the original spans) that should be removed from the results. We’ll then write a function to perform the filtering operation according to the specified list of UMLS codes, and display a new DataFrame that excludes all healthcare concepts connected to those codes.
# Variable containing a list UMLS codes from original `spans` to filter out
generic_concepts_list = ["UMLS:C0013227",
"UMLS:C0993159",
"UMLS:C0039225",
"UMLS:C0150618",
"UMLS:C0008679",
"UMLS:C0013182",
"UMLS:C0013182",
"UMLS:C0086418",
"UMLS:C4085337",
"UMLS:C0039798",
"UMLS:C0870814",
"UMLS:C1704938"
]
# function to filter out all UMLS codes listed above
# `scienceiodf` is the pandas df containing the `spans` from the `structure` endpoint
def remove_generic_concepts(scienceiodf,generic_concepts_list):
return scienceiodf[~(scienceiodf.concept_id).isin(generic_concepts_list)].reset_index(drop=True)
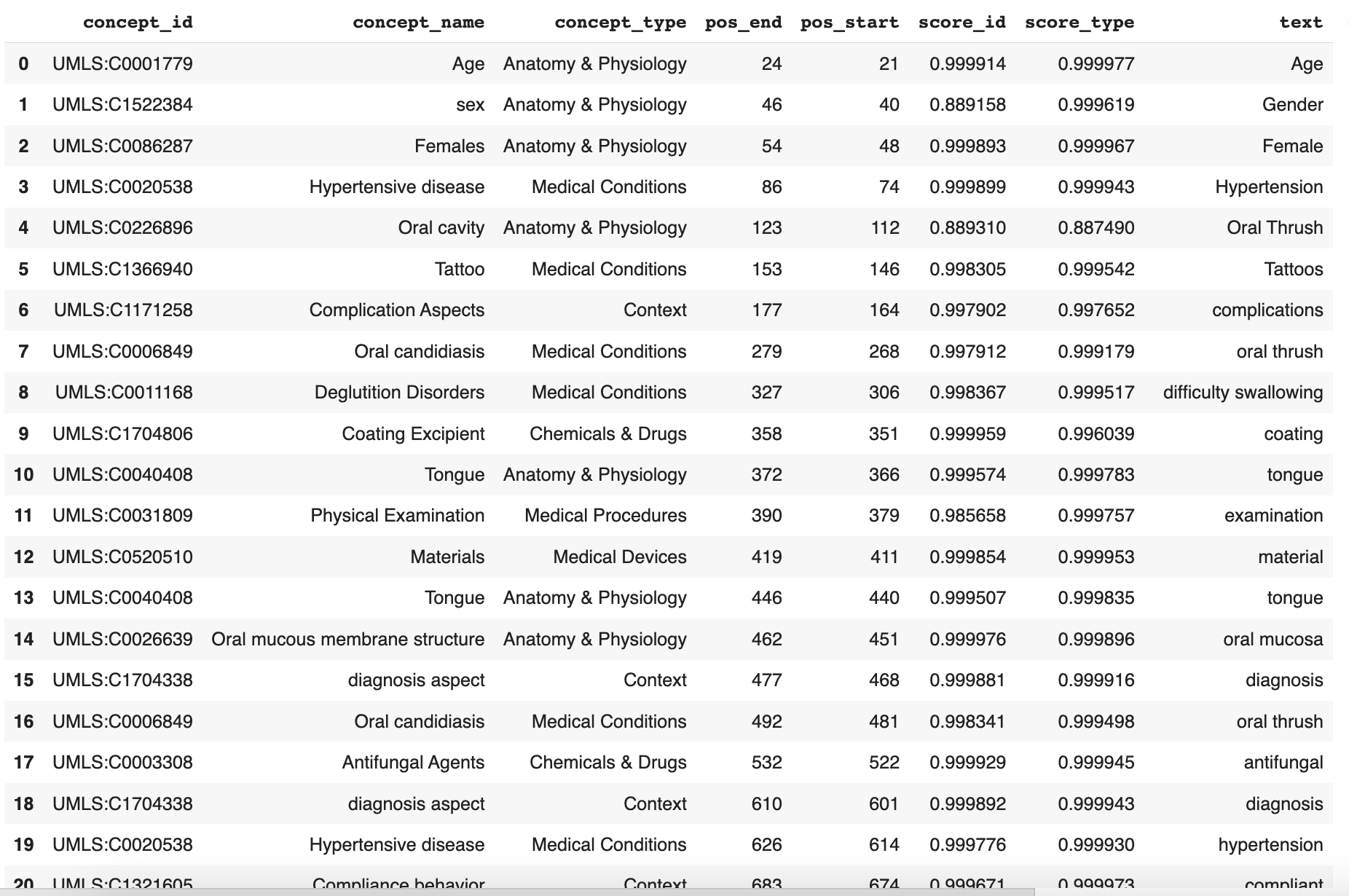
# Build a new df with `spans` containing all remaining UMLS codes
df = remove_generic_concepts(df,generic_concepts_list)
df.head(50)
The result looks like this (DataFrame truncated due to length; continues from line 20 and ends at line 37):
Option 2: Filter By Keyword
Use Case: I want to analyze all instances of one or more keywords/text strings, either within a specified concept or across the clinical note, and want to understand the context of each instance found.
This type of filtering might be used when looking for a patient record that mentions a specific keyword or concept, while also seeking to understand the contextual text around that keyword if it is found. Including the context as part of the filtering funtion allows us to quickly find exactly what we’re looking for without having to read the entire clinical note(s). It also allows us to quickly verify the accuracy of the model’s filtering behavior.
In this option, we’ll filter to find specific text strings in the clinical note regardless of where they fall in the original spans (in this case “oral”, “thrush”, and “hyperten”). To do this, we’ll first create a filter_concepts_show_neighborhood function that finds only the text strings we specify, renders the results in a new DataFrame, and includes contextual text around each text string found. Next, we’ll define the text strings in a filter_red_flagged_concepts variable and run the function.
'''
Function defining the filtering behavior and printing results
- `text` is the original text
- `scienceiodf` is the pandas df containing the original `spans`
- `filter_concepts` to only show results for text strings that match
- `neighborhood` to specify the length of contextual text shown
- `subsetdf` is the subsetted DataFrame of `scienceiodf`
'''
def filter_concepts_show_neighborhood(text: str,
scienceiodf: pd.DataFrame,
filter_concepts: List[str] =[""],
neighborhood: int = 50) -> pd.DataFrame:
filter_concepts = ("|").join([x.lower() for x in filter_concepts])
print("Looking for concepts that match the pattern:",filter_concepts)
scienceiodf = scienceiodf.copy()
subsetdf = scienceiodf.loc[scienceiodf["concept_name"].str.contains(
filter_concepts,case=False,na=False)].reset_index()
# print `subsetdf`
subsetdf["context_text"] = subsetdf.apply(lambda row:
text[max(0,row["pos_start"]-neighborhood):min(len(text),row["pos_end"]+neighborhood)],
axis=1)
print("Found ",subsetdf.shape,"concepts")
return subsetdf
# Specify words to include from the `subsetdf` here
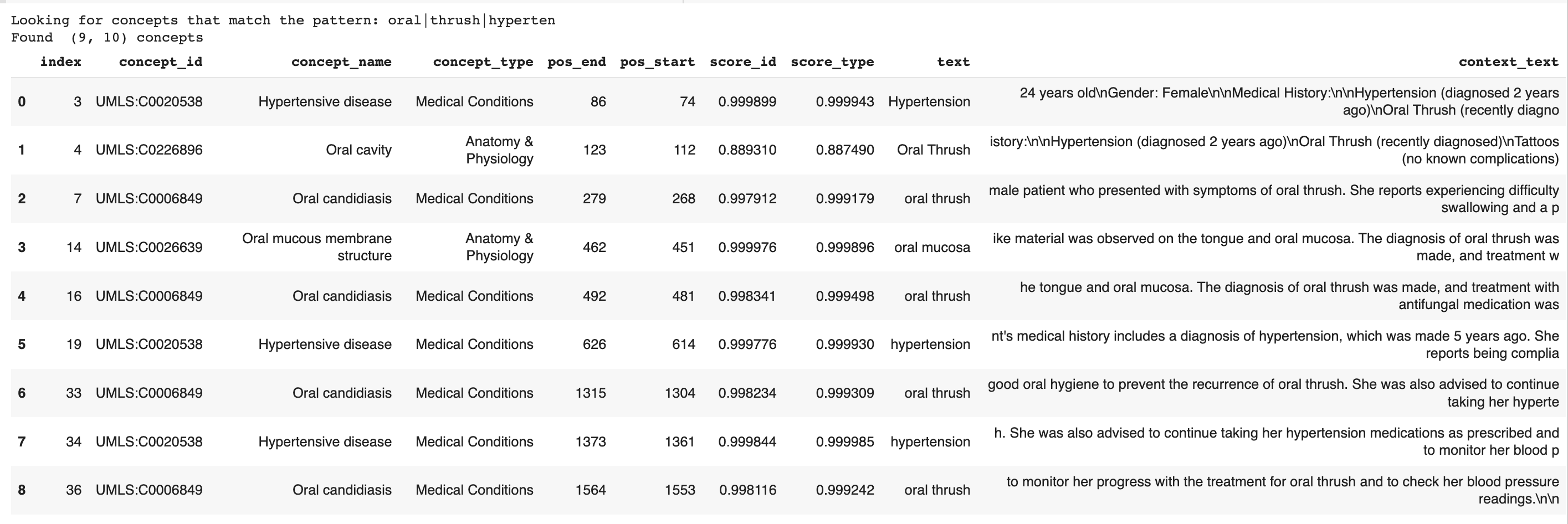
filter_red_flagged_concepts = ["oral","thrush","hyperten"]
filter_concepts_show_neighborhood(query_text, df, filter_red_flagged_concepts)
The result looks like this:
As another option, you could find only the text related to a specific value in concept_name (in this case, “tattoo”) by using the same function. Simply specify a different variable (filter_yellow_flagged_concepts) in the last two lines of the code.
'''
Function defining the filtering behavior and printing results
- `text` is the original text
- `scienceiodf` is the pandas df containing the original `spans`
- `filter_concepts` to only show results for text strings that match
- `neighborhood` to specify the length of contextual text shown
- `subsetdf` is the subsetted DataFrame of `scienceiodf`
'''
def filter_concepts_show_neighborhood(text: str,
scienceiodf: pd.DataFrame,
filter_concepts: List[str] =[""],
neighborhood: int = 50) -> pd.DataFrame:
filter_concepts = ("|").join([x.lower() for x in filter_concepts])
print("Looking for concepts that match the pattern:",filter_concepts)
scienceiodf = scienceiodf.copy()
subsetdf = scienceiodf.loc[scienceiodf["concept_name"].str.contains(
filter_concepts,case=False,na=False)].reset_index()
# print `subsetdf`
subsetdf["context_text"] = subsetdf.apply(lambda row:
text[max(0,row["pos_start"]-neighborhood):min(len(text),row["pos_end"]+neighborhood)],
axis=1)
print("Found ",subsetdf.shape,"concepts")
return subsetdf
# Specify the `concept_name` value to include from the `subsetdf` here
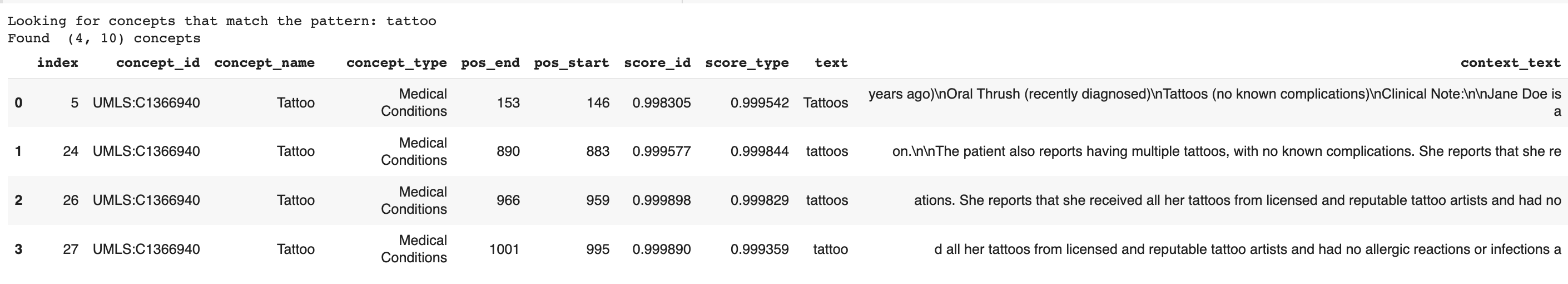
filter_yellow_flagged_concepts = ["tattoo"]
filter_concepts_show_neighborhood(query_text,df, filter_yellow_flagged_concepts)
The result looks like this:
Feedback
Was this page helpful?
Great! If you ever have questions or want to provide feedback, send us an email.
Bummer. We hate when we miss the mark. If you have suggestions for improvements or other general comments, send us an email.